Research Areas
Overview
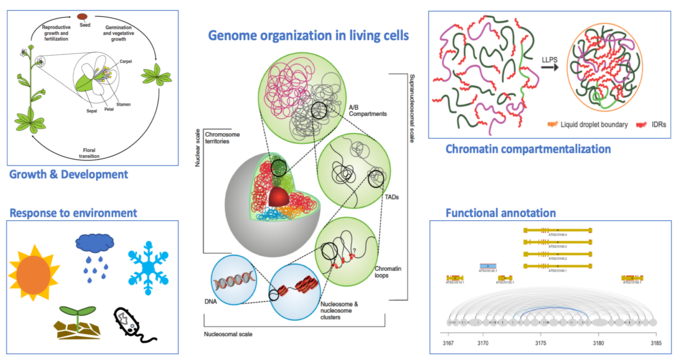
For most eukaryotes, their genomic DNA would be at least a few centimeters when fully stretched. Intriguingly, genomic DNA must be placed inside of a tiny nucleus with only a few micrometers in diameter. In the nucleus, what need to be properly coordinated in time and space, are various types of chromatin-related events such as transcription, DNA replication, recombination and repair, which ultimately determine how a cell behaves. It is well known that in the nucleus, DNA is not randomly packed; instead, it is organized hierarchically together with many proteins. How chromatin is packed, how the packing affects long-range chromatin interactions, and how the epigenomic and transcriptomic landscapes are connected to the 3D genome, are central questions in genome biology.
Large variations in chromatin structure are expected across the plant kingdom, as genomes among species are far more dynamic and plastic. At the moment, however, the functional and structural partition of chromatin domains, and the structural proteins regulating local chromatin packing in plants are poorly understood. In a long-term perspective, such knowledge is required for understanding miscellaneous chromatin activities in living cells and implementing structure-informed plant genome engineering towards generating varieties with desired and consistent agronomic traits.
Figure shown above: DNA sequences and epigenetic modifications, together with 3D chromatin organization, determines how a genome functions.
Three-dimensional chromatin organization
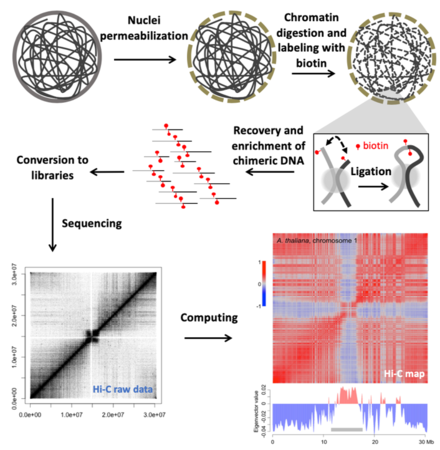
We focus on investigating the 3D-chromatin structure in plants, as well as its biological meaning in terms of transcriptional regulation in response to developmental cues and environmental changes. From a genome structure point of view, by combining high throughput sequencing, computing and modern molecular biology approaches, much of our work has been focused on integrative genome-wide analysis on the interplay among epigenetic marks, transcriptome, and chromatin organization. Nowadays, we are able to study chromatin structures in multiple plant species at an unprecedented resolution. We welcome talented and motivated people to further explore this emerging field with us.
Figure shown above: An overview of the Hi-C pipeline for investigating genome-wide chromatin interaction patterns. As the end product of this pipeline, the matrix (Hi-C map) tells us which parts in the genome are close amongst themselves. This matrix can also be analyzed by referring to many other chromatin features, such as transcription and epigenetic marks.
Chromatin positioning at the nuclear periphery
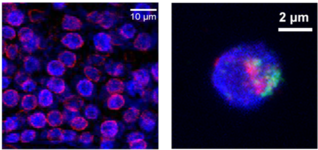
The nuclear space is not a homogeneous biochemical environment. A large number of studies in the animal field have demonstrated that the transcriptional activity of a gene is linked to its positioning in the nuclear space. Following the discovery of Lamina-Associated Domains (LADs) in animal genomes, which are transcriptionally repressed chromatin regions, the mechanisms regulating non-random positioning of chromatin at the nuclear periphery and their biological relevance have been intensively studied. Nowadays, it is known that animal lamin proteins play essential roles in anchoring LADs to the nuclear periphery. However, plants do not have lamin proteins; how and why plants exhibit selective chromatin positioning at the nuclear periphery are poorly understood.
Recently, we have discovered that some plant-specific proteins function as “lamins”, which are required in regulating chromatin organization at the nuclear periphery in plants. At the moment, we are working on revealing molecular mechanisms of how plant “lamins” function to regulate plants’ growth and development.
Figure shown above: Left: Specific localization of plant “lamin” proteins (red) at the nuclear border. Right: Differential positioning of chromatin (red and green) at the nuclear periphery.